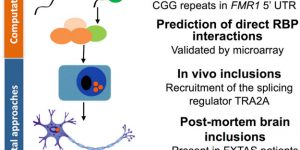
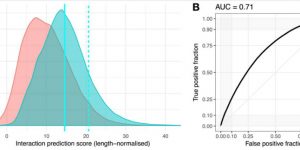
Cid-Samper F, Gelabert-Baldrich M, Lang B, Lorenzo-Gotor N, Ponti RD, Severijnen LWFM, Bolognesi B, Gelpi E, Hukema RK, Botta-Orfila T, Tartaglia GG.
“An Integrative Study of Protein-RNA Condensates Identifies Scaffolding RNAs and Reveals Players in Fragile X-Associated Tremor/Ataxia Syndrome.”
Cell Rep, 25(12):3422-3434.e7, doi: 10.1016/j.celrep.2018.11.076, 2018.
Delli Ponti R, Armaos A, Marti S, Tartaglia GG.
“A Method for RNA Structure Prediction Shows Evidence for Structure in lncRNAs.”
Front Mol Biosci, 5:111, doi: 10.3389/fmolb.2018.00111, 2018.
Miotto M, Olimpieri PP, Di Rienzo L, Ambrosetti F, Corsi P, Lepore R, Tartaglia GG (corresponding author), Milanetti E.
“Insights on protein thermal stability: a graph representation of molecular interactions.”
Bioinformatics, doi: 10.1093/bioinformatics/bty1011, 2018.
Lang B, Armaos A, Tartaglia GG.
“RNAct: Protein-RNA interaction predictions for model organisms with supporting experimental data.”
Nucleic Acids Res, doi: 10.1093/nar/gky967, 2018.
Qamar S, Wang G, Randle SJ, […], Tartaglia GG,[…], St George-Hyslop P.
“FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions.”
Cell, 173(3):720-734, 2018.